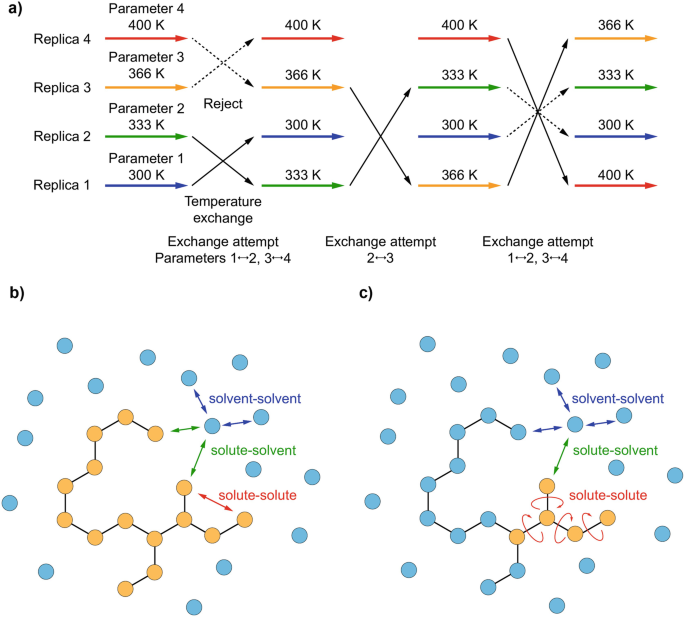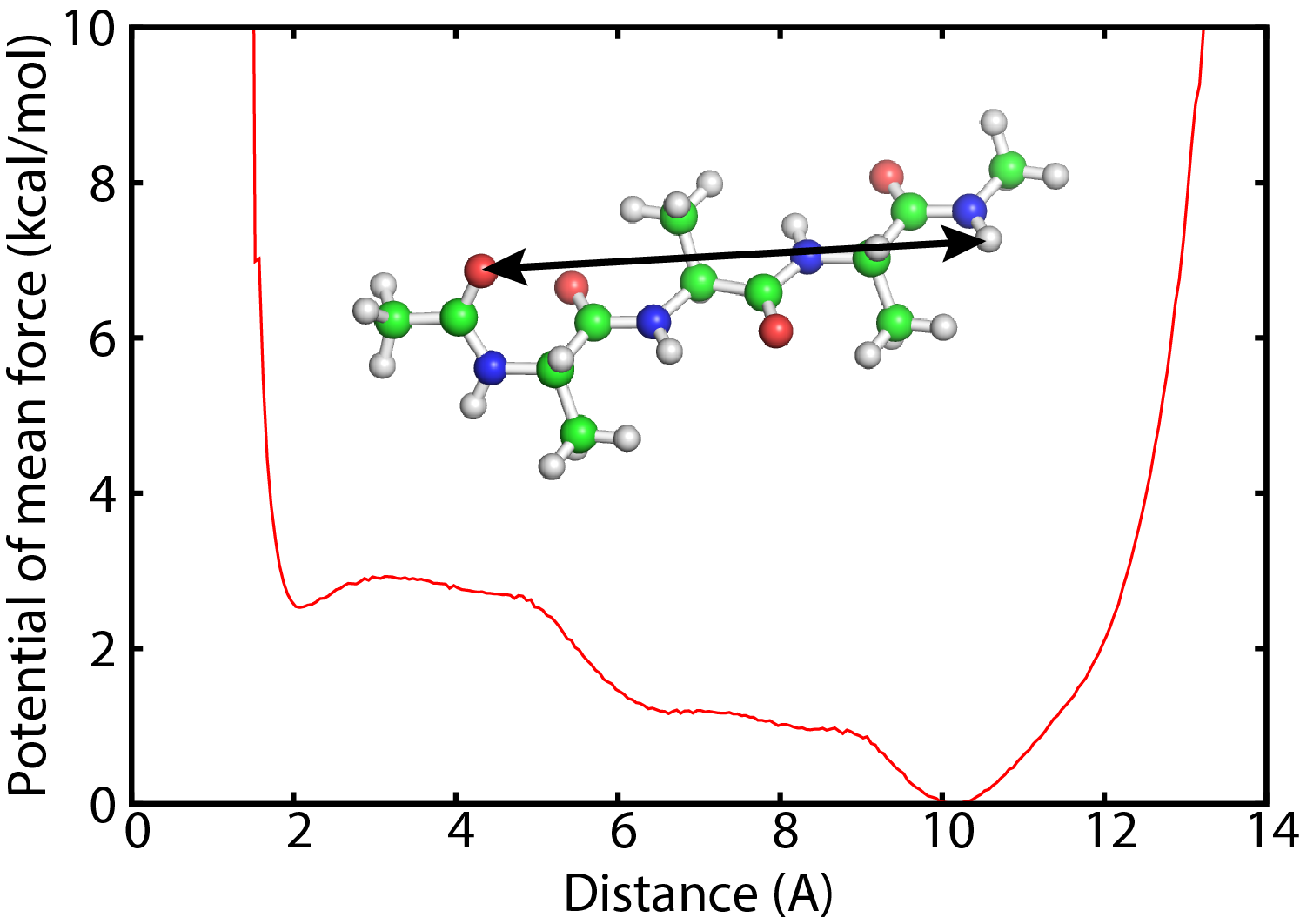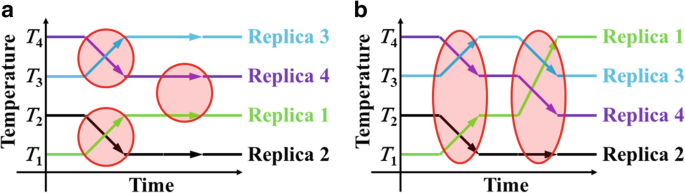
All-Atom Molecular Dynamics Simulation Methods for the Aggregation of Protein and Peptides: Replica Exchange/Permutation and Nonequilibrium Simulations | SpringerLink
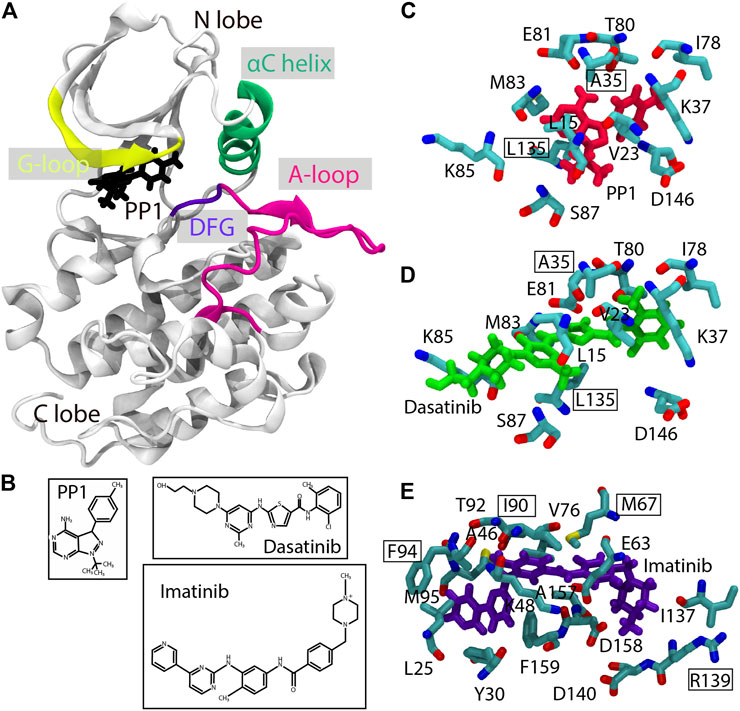
Frontiers | Practical Protocols for Efficient Sampling of Kinase-Inhibitor Binding Pathways Using Two-Dimensional Replica-Exchange Molecular Dynamics

Kinetics from Replica Exchange Molecular Dynamics Simulations | Journal of Chemical Theory and Computation

Replica exchange with solute tempering: A method for sampling biological systems in explicit water | PNAS

A two-dimensional replica-exchange molecular dynamics method for simulating RNA folding using sparse experimental restraints - ScienceDirect
Accelerated flexible protein-ligand docking using Hamiltonian replica exchange with a repulsive biasing potential | PLOS ONE

IJMS | Free Full-Text | A Hamiltonian Replica Exchange Molecular Dynamics ( MD) Method for the Study of Folding, Based on the Analysis of the Stabilization Determinants of Proteins
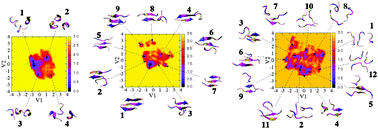
Effects of all-atom force fields on amyloid oligomerization: replica exchange molecular dynamics simulations of the Aβ16–22 dimer and trimer - Physical Chemistry Chemical Physics (RSC Publishing)

Molecular Dynamics Simulations Using Temperature-Enhanced Essential Dynamics Replica Exchange: Biophysical Journal

Molecular dynamics simulations of biological membranes and membrane proteins using enhanced conformational sampling algorithms - ScienceDirect

Increasing the Sampling Efficiency of Protein Conformational Change by Combining a Modified Replica Exchange Molecular Dynamics and Normal Mode Analysis | Journal of Chemical Theory and Computation

Exploring Multiple Binding Modes Using Confined Replica Exchange Molecular Dynamics. | Semantic Scholar
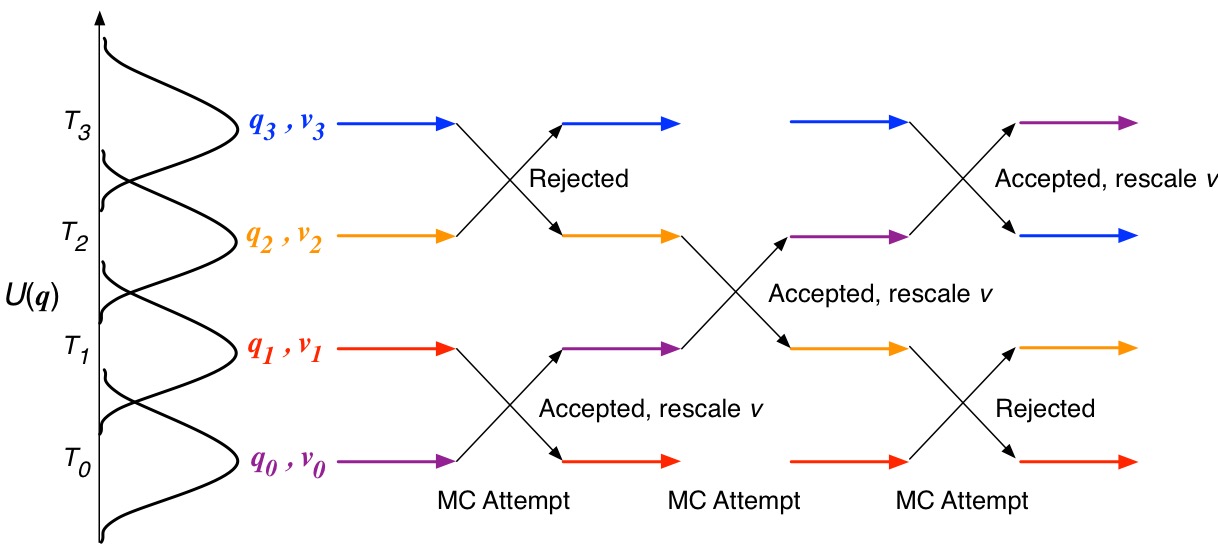
Mass-Scaling Simulated Tempering for Optimizing Time-steps - Scratchings on biomolecular simulation and theory

Replica Exchange Molecular Dynamics: A Practical Application Protocol with Solutions to Common Problems and a Peptide Aggregation and Self-Assembly Example. - Abstract - Europe PMC